Pericentromeric hSATII RNA changes chromosomal interaction via CTCF... | Download Scientific Diagram

TET‐catalyzed oxidation of intragenic 5‐methylcytosine regulates CTCF‐dependent alternative splicing | The EMBO Journal
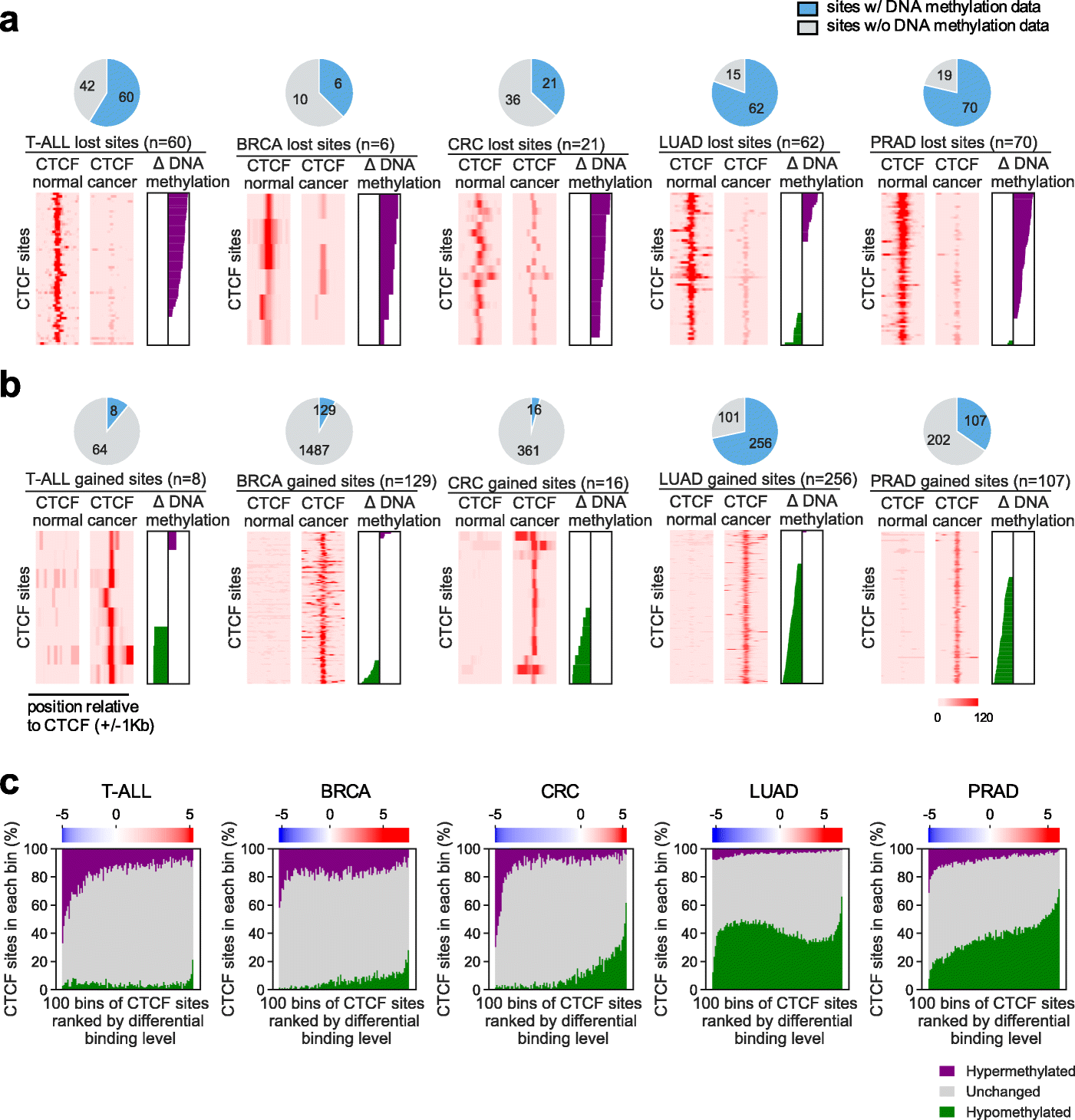
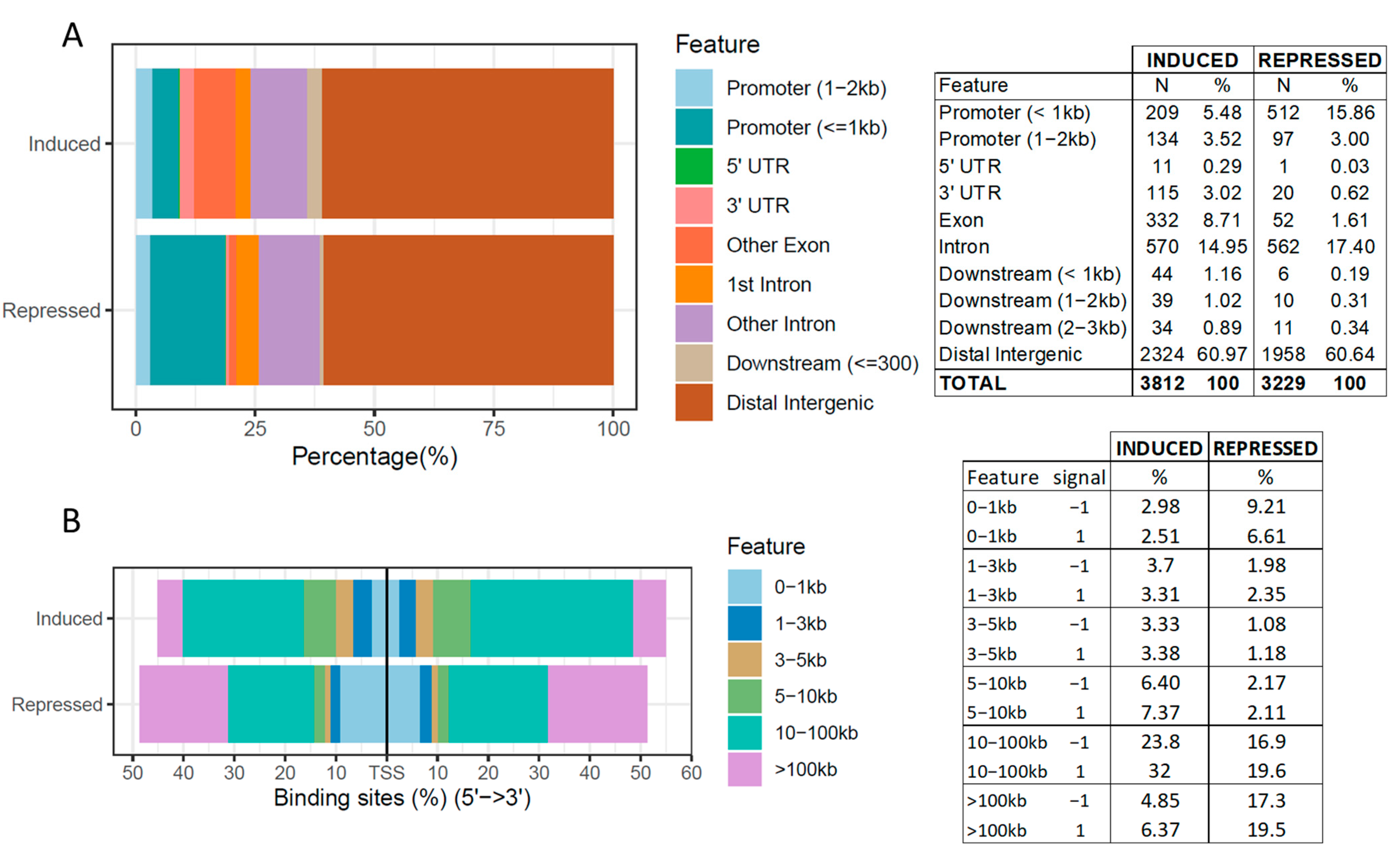
Cancer-specific CTCF binding facilitates oncogenic transcriptional dysregulation | Genome Biology | Full Text

Cell-specific exon methylation and CTCF binding in neurons regulates calcium ion channel splicing and function | bioRxiv

Locus-Specific Targeting to the X Chromosome Revealed by the RNA Interactome of CTCF - ScienceDirect

Constitutively bound CTCF sites maintain 3D chromatin architecture and long-range epigenetically regulated domains | Nature Communications

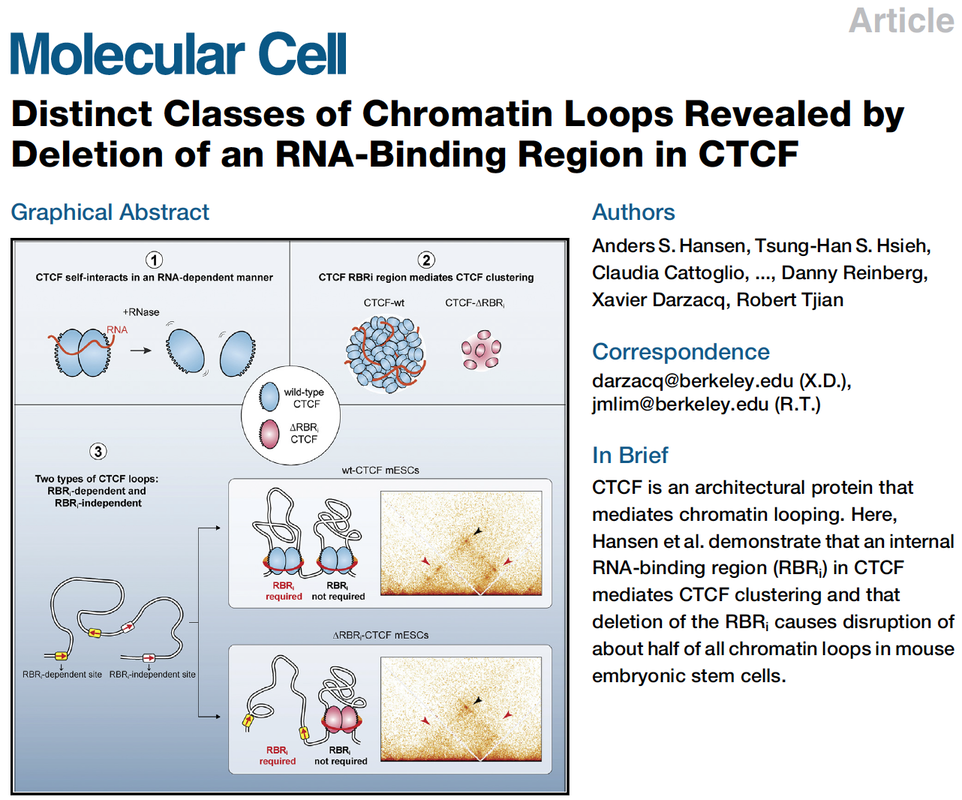
Identification and characterization of the CTCF RBR. (A,C) Schematic... | Download Scientific Diagram

DNA Methylation Regulates Alternative Polyadenylation via CTCF and the Cohesin Complex - ScienceDirect

Genome-wide Studies of CCCTC-binding Factor (CTCF) and Cohesin Provide Insight into Chromatin Structure and Regulation* - Journal of Biological Chemistry

Molecular mechanism of directional CTCF recognition of a diverse range of genomic sites | Cell Research

CTCF: the protein, the binding partners, the binding sites and their chromatin loops | Philosophical Transactions of the Royal Society B: Biological Sciences

CTCF-Mediated Enhancer-Promoter Interaction Is a Critical Regulator of Cell-to-Cell Variation of Gene Expression - ScienceDirect